Study population
From each CT volume, we automatically identified the slice at L3 using a previously published convolutional neural network (CNN) algorithm25, manually verifying correctness for each case. The L3 slice was chosen as it is the most common reference location for BC analysis26,27,28,29. The process to select the final cohorts of patients is shown in Supplementary Fig. 1. We excluded images with artifacts that obscured the L3 level (e.g., spinal instrumentation), those that had anatomical variations in the image (e.g., scoliosis) that precluded the assignment of a single slice to the L3 level, those that did not contain the L3 slice in the field of view, and those obtained within the same 6-month window as an already included image. For each cohort, we used random sampling (stratifying on outcome labels) to divide patients in the dataset into training and test datasets representing 80% and 20% of the images, respectively, for IHD risk estimation and model creation.
Segmentation model
Given that BC metrics from manual segmentations have been correlated with cardiovascular risk, we trained a 2.5D U-Net CNN to perform BC analysis for segmenting regions of muscle along with VAT and SAT on an abdominopelvic CT slice30. A total of 400 axial L3 slices obtained exclusively from the training set and manually labeled were used during model tuning and evaluation. Manual segmentation of muscle, VAT, SAT, and bone were performed semi-automatically with CoreSlicer31, a free online tool, using attenuation thresholds and manual adjustments as needed (AW, 5 years of experience). 320 images were randomly selected for segmentation training, 40 for validation, and 40 for testing. Segmentation performance on the 40 test images was determined using segmentation accuracy metrics, namely the Dice coefficient and root-mean-squared coefficient-of-variation. The Dice coefficient is calculated as two times the ratio of the intersection between the ground-truth and segmented image masks to the sum of the number of pixels in each mask. A Dice score of 1 indicates a perfect segmentation. The variations between the manual and automated segmentation approaches on the tissue-wise radiodensity (measured in Hounsfield units [HU]) and cross-sectional area (measured in cm2) were also evaluated.
The inputs to the 2.5D network were individual 2D axial CT slices at the L3 level with three different window and level (W/L) settings for maximizing tissue contrast. The W/L settings that were used included a soft tissue window (W/L = 400/50 HU), a bone window (W/L = 1800/400 HU), and a custom window (W/L = 500/50 HU). After applying the appropriate windowing, each of the channels was normalized to values between 0 and 1.
The U-Net utilized 6 convolution levels (each with two convolutional operators, both followed by a rectified linear unit activation, followed by batch normalization) for the encoder and decoder32. The number of U-Net features per layer increased quadratically from 32 to 1024. The dimensions of the convolutional kernels were 3 × 3, while that of the maximum pooling operator was 2 × 2. A softmax activation was used as the final layer in the CNN along with a weighted soft Dice loss function to account for class imbalance amongst the segmented tissues. The U-Net hyperparameters had previously been optimized for medical imaging segmentation32. A weighting factor of 8 was used for muscle during loss computation. All network weights were randomly initialized using the He et al.33 initialization scheme.
Training was performed with the Adam optimizer with default parameters β1: 0.9, β2: 0.999, with a learning rate schedule that included a base learning rate of 1e−3 and the learning rate being reduced by 0.8 for every epoch to a minimum value of 1e−8. The network was nominally trained for 130 epochs with an early stopping criterion of a minimum change in loss of 1e−5 and a patience of 8 epochs. The batch sizes for training, validation, and testing were chosen to be 10, 33, and 80, respectively, for maximizing GPU memory. Training was performed using a Tensorflow 1.14 on an NVIDIA Titan Xp GPU.
We used the segmentations generated by the U-Net model and determined average muscle radiodensity in HU and the VAT/SAT cross-sectional area ratio. We trained a model (L2 logistic regression) using tenfold cross validation to select the L2 penalty weight. The model was trained using the training sets to predict IHD outcome at 1 and 5 years using these two features. We refer to this as the Segmentation Only model.
Imaging only model
We trained a CNN as a feature extractor to predict the risk of IHD using a single axial slice at the L3 level, using an EfficientNet-B6 architecture34. EfficientNets were designed to balance the scaling of network width, depth, and image size, thus producing state-of-the-art results in conventional image classification tasks with smaller and faster models as compared to traditionally used feature extractors, such as ResNet5035. The 512 × 512 pixel grayscale L3 slice was clipped to contain values from − 1000 to 1000 HU, represented as an unsigned integer, replicated thrice to produce a 3 × 512 × 512 image, and resized to 3 × 528 × 528 to be input into the network. The initial EfficientNet-B6 model weights were obtained from a pre-trained model optimized for ImageNet classification performance (https://pypi.org/project/efficientnet-pytorch/)36. The final fully-connected layer was replaced with one corresponding to a binary outcome, and the model weights were fine tuned. The tuning of the weights was performed with a cross-entropy objective using a random selection of 80% of the training set, reserving 20% for validation. Training was performed for multiple epochs until no improvement in validation loss or Area Under the Receiver Operating Characteristic (AUROC) was observed. A batch size of 8 and an Adam optimizer37 was used with default parameters β1 0.9 β2 of 0.999 and a constant learning rate of 7e-6 and 6e-6 for the 1 and 5-year cohorts, respectively. Model training was carried out using Pytorch 1.1 on an NVIDIA Titan Xp GPU.
We compared training only the final layer as opposed to training all of the model weights, using additional image augmentations such as rotations of up to 3° and pixel shifting of up to 5 pixels during training, and using a focal loss function assigning higher weights to IHD cases. We chose the final network architecture and training strategy described above as it achieved the highest AUROC in the validation stage (Supplementary Table 1).
Clinical only model
To avoid sparsity, we grouped ICD10, CPT, and medications based on their underlying ontology. Namely, we grouped ICD10 codes (https://bioportal.bioontology.org/ontologies/ICD10) by blocks and CPT Category I codes (https://bioportal.bioontology.org/ontologies/CPT) at the H2 level. To further summarize overall burden of disease in a single feature, we also calculated the Charlson Comorbidity Index for each patient and included it as a feature39. Irrespective of dose and frequency, the active substance of prescribed medications was mapped to RxNorm and subsequently to the second level of the Anatomical Therapeutic Code (https://bioportal.bioontology.org/ontologies/ATC), corresponding to the therapeutic subgroup. Furthermore, prior to training, we iteratively removed highly correlated features (Pearson correlation coefficient > 0.5). In all, each patient EMR was represented using a 422-dimensional vector. The final clinical features used and their descriptions are listed in Supplementary Table 2.
The predictive model used for predicting IHD risk from EMR features was designed using XGBoost, an optimized gradient-boosting machine learning system40. In gradient boosting, an ensemble of weak learners is iteratively constructed by greedily adding estimators that fit the previous residual. In doing so, gradient boosting algorithms can perform successfully across a wide variety of predictive tasks, often outperforming traditional models such as logistic regression or support vector machines. We chose XGBoost for its robust performance in predictive modeling, and for its capacity to handle missing data, which other gradient boosted methods like AdaBoost lack. Optimal parameters for training the model were established using ten-fold cross-validation on the training set.
Fusion models
To further identify the potential benefits of using imaging BC features as predictors of IHD risk, we constructed three models to fuse imaging and clinical data. In the first fusion, we concatenated the features used by the PCE with the average muscle radiodensity and the VAT/SAT ratio (PCE + Segmentation Model), the latter two measurements obtained by using our automated segmentation model. We used an XGBoost classifier to predict IHD risk with the PCE + Segmentation features. In the second fusion, we combined the risk output from our EfficientNet-B6 model with the risk output from our medical record model using stacking with L2 logistic regression (Imaging + Clinical Model). In the third fusion, we combined the risk output from the Imaging Only, Clinical Only, and Segmentation Only models (Imaging + Clinical + Segmentation Model) in a stacking L2 logistic regression classifier. In all fusion cases, we performed a hyperparameter search in tenfold cross-validation in the training set. A summary of our prediction model approach can be seen in Fig. 1. The clinical model and fusion models were trained using scikit-learn 0.23 (https://scikit-learn.org/) in Python 3.6.
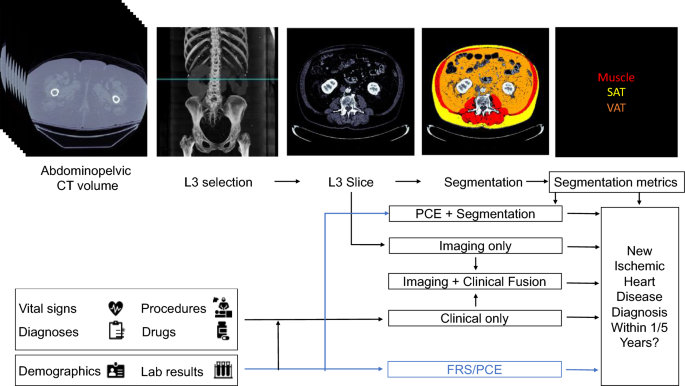
Proposed models for evaluating risk of a future ischemic heart disease diagnosis in one or five years following an abdominopelvic computed tomography (CT). The blue line shows which sources are used by existing risk assessment models, the Framingham Risk Score (FRS) and Pooled Cohort Equations (PCE). The PCE is the standard tool for atherosclerotic cardiovascular disease risk assessment in current clinical guidelines for 10-year risk estimation. In our proposed models, the axial slice corresponding to the third lumbar vertebra anatomical level (L3) is automatically selected from the CT volume. In one model, the L3 slice is automatically segmented to extract mean muscle radiodensity in Hounsfield units and the Visceral/Adipose cross-sectional area ratio; these features are used alone or in combination with covariates from the PCE to form a Segmentation or PCE + Segmentation Model. Alternatively, features are automatically extracted from the L3 slice using a convolutional neural network (Imaging Only Model). As an additional approach, predictions from a model trained with features constructed from the patient’s electronic medical record within the year prior to CT acquisition (Clinical Only Model) are stacked with those of the imaging model (Imaging + Clinical Fusion Model) and with those of the Segmentation model (Imaging + Clinical + Segmentation Model, not depicted).
Interpretation of model performance
Two baseline models currently employed in clinical practice that estimate 10-year cardiovascular risk were used as a reference, namely the FRS1 and the PCE2. We studied the performance of the FRS as it directly models the risk of hard IHD events. Despite the PCE including other atherosclerotic cardiovascular disease outcomes, we also included them as a baseline given their current use in clinical practice guidelines. Since several patients were missing covariates necessary for FRS and PCE calculation (Supplementary Table 3), these values were imputed using median imputation to allow for a baseline risk calculation for all individuals in the study. In addition, we examined the performance of all models in the subpopulations with available/missing PCE covariates (Supplementary Table 6).
Attribution analysis
With the aim of allowing for interpretability of the fusion model, we examined the contributions of individual features in both the imaging and clinical models.
For the imaging model, we developed a new tissue saliency interpretation tool as described in the introduction to evaluate the tissues that had a large contribution to the final prediction outcome. We first calculated the derivative, \(w\), of the IHD class score at the final layer of our EfficientNet-B6 model, \({S}_{{\text{IHD}}}\), with respect to each input pixel, \({I}_{ij}\), in the image \(I\)41. That is,
$${w}_{ij}= {\left.\frac{\partial {S}_{{\text{IHD}}}}{\partial I}\right|}_{{I}_{ij}}.$$
Using our segmentation model, each pixel was assigned a specific tissue class, \(t\). In our particular case, this corresponded to either muscle, VAT, SAT, other body tissues, or background. We obtain the observed normalized tissue saliency, \({S}^{O}\), for a particular tissue, as:
$${S}_{t}^{O}=\frac{|{w}_{ij}
Source link


