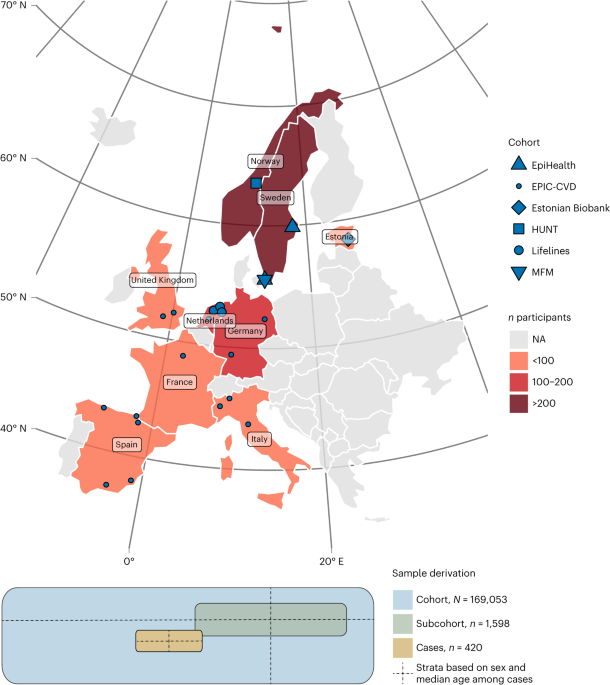
Study sample and results
The MIMI study sample draws biobanked blood and data from six European cohorts of the BBMRI-LPC (Biobanking and Biomolecular Research Infrastructure – Large Prospective Cohorts) collaboration.twenty two, as shown in Figure 1 and Supplementary Table 1. After determining the sample size, we provided each cohort with a standardized protocol (all definitions explained in detail) and an R script to select cohort representatives for subcohorts (Supplementary Note).
Cohort participants with a biobanked sample (at least 250 μl of plasma or serum, ultimately containing only plasma) and no previous clinical cardiovascular disease were eligible for inclusion in the study . Exclusion criteria were the presence before baseline of any of the following clinical cardiovascular diseases (myocardial infarction, coronary artery surgery, heart failure, structural heart disease, tachyarrhythmia, stroke, thromboembolic disease and peripheral vascular disease). (as defined) and kidney disease. Failure.
IMI identifies individuals whose primary cause of hospitalization or death within 6 months of baseline is acute myocardial infarction (International Classification of Diseases, 10th edition (ICD-10), I21, ICD-9, 410.0-410.6 and 410.8). defined as a case. We included both ST-elevation myocardial infarction and non-ST-elevation myocardial infarction. We encouraged efforts to include only type 1 myocardial infarction by not counting cases with any of the following ICD codes in a secondary position: anemia (ICD-10, D50 to D64, ICD -9, 280 to 285, etc.), tachyarrhythmia (ICD-10, D50 to D64, ICD-9, 280 to 285, etc.), e.g. ICD-10, I47 to I49, ICD-9, 427), heart failure ( (e.g., ICD-10, I50, ICD-9, 428), renal failure (e.g., ICD-10, N17 to N19, ICD-9, 428), chronic obstructive pulmonary disease (ICD-9, 584-586), -10, J43-J44, ICD-9, 491, 492, 496, etc.), sepsis and other serious infections (ICD-10, A40, etc. -A41; ICD-9, 038), or hypertensive crisis .
exposure
All blood samples were randomly divided into appropriate measurement plates, stratified by cohort (with similar numbers from each cohort on each plate), and aliquoted into plates. Quality control is summarized below and explained in detail in the supplementary notes.
Protein measurements were performed using the Olink proximity extension assay (Olink), a highly specific 92-plex immunoassay. Overall, 829 proteins across nine panels (cardiometabolic, cardiovascular II, cardiovascular III, development, immune response, inflammation, metabolism, tumor II, and organ damage) were analyzed. It contains 804 unique proteins (accounting for overlap between panels).Relative protein value on log2 The scale is reported with each protein value normalized per plate by centering all plates at the same median value, assuming random plate placement. Values below the assay limit of detection (LOD) were also included in the analysis.
Metabolites were analyzed using the UPLC–tandem MS (UPLC–MS/MS)-based Metabolon platform (Metabolon) in four different ways: -phase UPLC-MS/MS using negative mode electrospray ionization, and hydrophilic interaction LC/UPLC-MS/MS using negative mode electrospray ionization. Overall, 1,135 metabolites were captured, of which 925 were of known identity and 210 were of unknown identity. Relative metabolite levels were determined and normalized by date of analysis.Metabolite levels were log2 converted to undetectable level (
Samples that did not meet quality control criteria were initially excluded. Exclusion filters were applied separately to proteomics and metabolomics analyses, and only samples that passed quality control for both analyzes were included in the analysis set. For proteomic analysis, samples where more than 50% of the panel failed for technical reasons were excluded (n Exclude = 33). For metabolomics analysis, samples were excluded due to low volume or detection of fewer metabolites than expected (n Exclude = 4). As a result, a sample of 420 cases and 1,598 subcohort representatives remained for analysis.
We then used the same exclusion filters for proteins and metabolites to exclude biomarkers with very high proportions of undetectable or below LOD measurements. Biomarkers had to be detected in all six cohorts with at least 30 detectable values across all cohorts (approximately 1.5% of MIMI samples), otherwise they were excluded. As a result, 817 proteins (with some overlap) and 1,025 metabolites were retained for analysis.
statistical analysis
All analyzes were performed using R (version 4.1.1).twenty four Using GLM Nettwenty five,mouse26,rms27troops28 and survival29 Add-on package.
Analysis of each cause
In the discovery sample, associations of all clinical variables (listed in Extended Data Table 1), proteins, and metabolites with IMI were individually weighted with adjustment for covariates, as described below. It was analyzed with a stratified Cox proportional hazards regression model. Inverse sampling probability weights (Borgan II) are applied to account for the case-cohort design of the stratified model, accounting for different shapes of the baseline hazard for each MIMI cohort (6 levels), and a robust variance estimator (Huber- White) was used. ). Nonlinear relationships between continuous covariates (not including biomarkers) and IMI were modeled using restricted cubic splines, and all factor variables were considered unordered.
Associations with an FDR (Benjamini-Hochberg) of <0.05 were carried forward into the validation sample, yielding directionally consistent results. P < 0.05 is considered repeated.
Missing and sensitivity analysis
Clinical variables with high levels of missingness (previous smoking exposure, alcohol intake, physical activity) were not used in the analysis. Protein values below the LOD were included in the analysis. Undetectable metabolite levels were replaced with constant values and missing indicators were added, as described below. Remaining missing values for covariates were multiple imputed (nimputation = 20) using chained equations that include the outcome, clinical covariates, and other variables that are correlated with the variables in the imputation model.30.Regression results across the imputed dataset were combined using Rubin’s rule31.
The following secondary sensitivity analyzes were included: random effects inverse variance weighted meta-analysis (DerSimonian-Laird) combining outcomes by cohort, leave-one-out analysis examining single cohort effects, and imputing missing values. We do not analyze clinical covariates in a complete case analysis and limit the follow-up period to 3 months.
Perform modeling and predictive model development simultaneously
To assess whether biomarkers added to the clinical prediction model improve risk prediction, linear predictors from the prediction model were used as offsets in the LASSO Cox regression model. All proteins and metabolites were adjusted for technical variables before fitting the model. Briefly, each biomarker was used as an outcome variable in a regression model with all technical variables as covariates. The residuals from these models were used in place of the original biomarker values in the LASSO model. The fit of the LASSO model was bootstrapped 250 times to investigate the stability of variable selection.
Because biomarkers have nonlinear associations with outcomes and can interact, and because we have little prior knowledge of nonlinearities and interactions between these variables, a random forest of 2,000 trees was used as an exploratory analysis. fitted to the data. Simply put, a random forest adapts survival trees to bootstrap data samples using a random subset of variables within each tree, naturally handling interactions and nonlinearities. A variable importance measure is associated with each variable and is calculated based on the number of folds in which the variable participates. The random forest was bootstrapped 250 times to obtain CIs for variable importance measures.
Further analysis of relevant biomarkers
consent
This study was approved by the Uppsala Ethics Authority (Dnr 2016/197). All Estonian Biobank participants signed an extensive informed consent form. This study was carried out under ethics approval 258/M-21 from the Research Ethics Committee of the University of Tartu and data release J08 from the Estonian Biobank. The Lifeline protocol was approved by the Medical Ethics Committee of the University Medical Center Groningen with number 2007/152. This study was conducted in accordance with the Declaration of Helsinki. The EpiHealth study was approved by the Ethics Committee of Uppsala University, and all participants provided written informed consent. MFM was approved by the previous Regional Research Committee (2014/643) in Lund, Sweden, and all participants provided informed consent. The EPIC-CVD cohort’s institutional review board approved the study protocol, and all participants provided written informed consent. Participation in the HUNT study was based on informed consent, and the Data Inspectorate and the Norwegian Regional Ethics Committee for Medical Research approved the study.
Report overview
For more information on the study design, please see the Nature Portfolio Reporting Summary linked in this article.